Detection of antigen-specific immunity (Cellular Immunology)
Mass spectrometry-based immunopeptidomics for sensitive HLA-peptide detection
T cell interactions with HLA presented peptides lie at the heart of any immune response. TIGL has developed and provides:
- Serial HLA-I and HLA-II isolation from a single sample
- Single-allele systems to characterize the rules of HLA peptide processing and presentation
- Proteogenomic approaches to generate patient-specific protein databases for antigen discovery
- Direction detection of multiple antigen classes, including TAAs, CTAs, neoantigens, ERVs, and non-canonical peptides
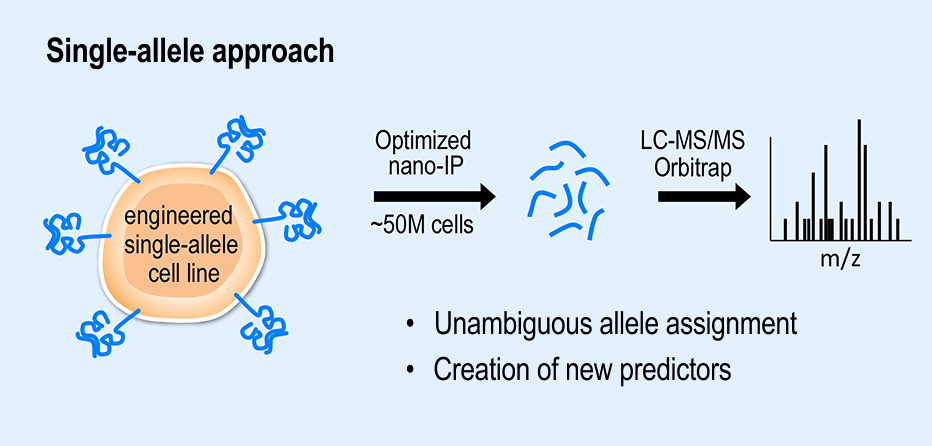
In our single-allele approach, B721.221 cells (~50 M) are transduced to express only one HLA allele. Immunopurified peptides are analyzed by LC-MS/MS and sequenced via an HLA-allele-specific database search (Abelin, Keskin et al Immunity 2017)
Immune-monitoring
The strength of TCR binding to cognate antigen (functional avidity) is strongly correlated with in vivo T cell effector function. TIGL has developed and provides:
- ELISPOT assays and multicolor flow cytometry to investigate antigen-specific T cell responses
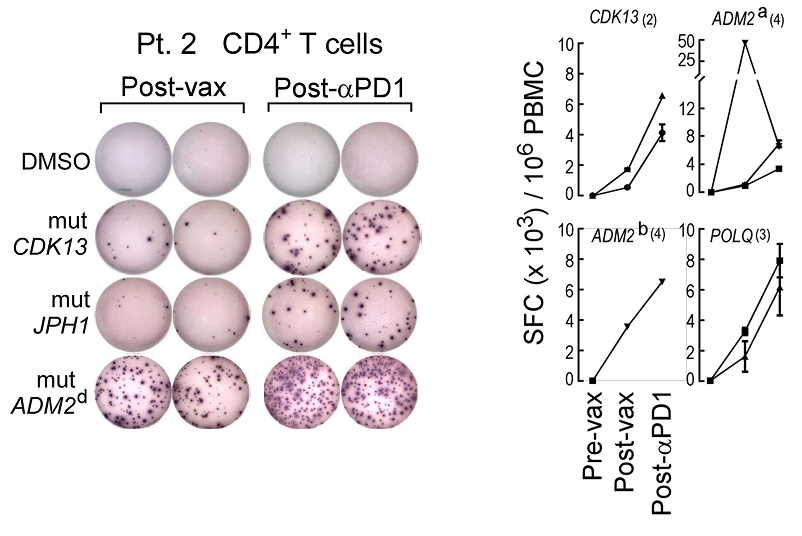
Example of IFN-γ ELISPOT results are shown for CD4+ T cells specific for mutated CDK13, JPH1, and ADM2d at week 16 after vaccination and after PD-1 blockade for patient 2. Complete response kinetics to mutated peptides are also shown (Ott and Hu et al Nature 2017).
TCR reconstruction
TIGL has developed and provides:
- A novel TCR reconstruction pipeline that enables the testing of neoantigen-specific TCRs for functional avidity as an indicator of anti-tumor activity using ELISPOT and intracellular cytokine staining
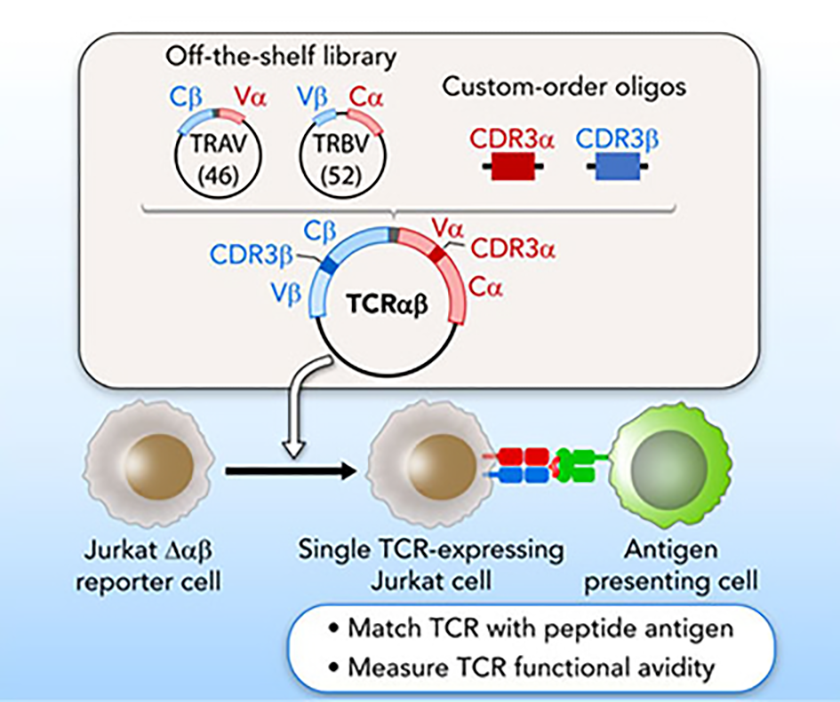
A streamlined approach for identifying and reconstructing TCRs, and testing for specificity to candidate antigens (Hu, Anandappa et al Blood 2018).
Staff
Derin B. Keskin, PhD, Immunology Lead Scientist
Katarina Pinjusic, PhD, Postdoctoral Fellow
Allison Vanasse, Research Technician
Joseph Duke-Cohan, Research Technician, Bioinformatics
Melis Akinci, Research Technician
Marwa Belhaj, Research Technician