Immunogenomic Analysis
The interdependence of immunology and cancer biology has been strengthened with recent advances in immunogenomic analysis through computational approaches.
We aim to take advantage of the data generated from high-throughput sequencing technologies to systematically identify neoantigens across cancers, antigen presentation states, biomarkers of response, and genetic and epigenetic states of the tumor and host.
TIGL has developed and provides:
- Bulk transcriptome analysis
- Single-cell transcriptome analysis
- TCR analysis
- Methylation state and RBS detection
- Neoantigen prediction
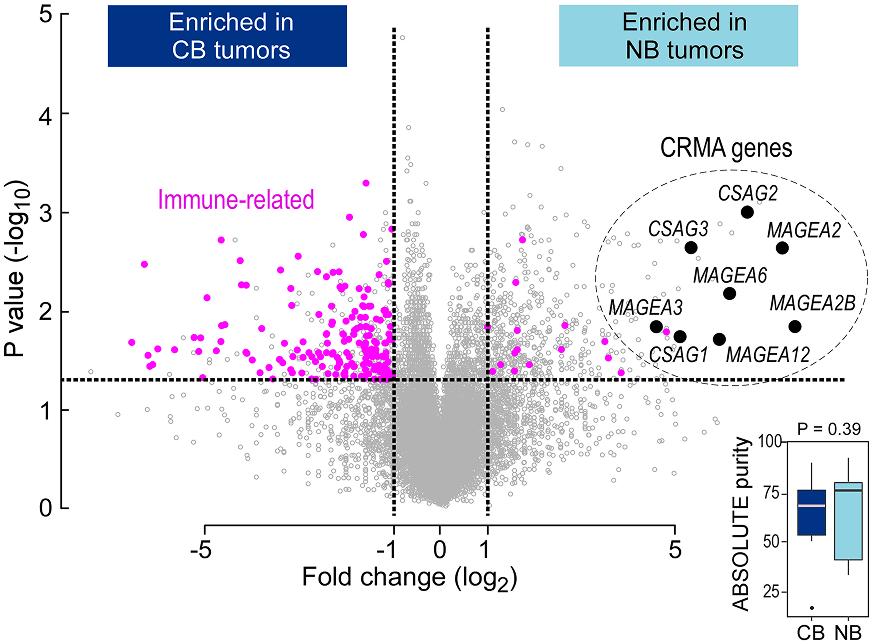
A differential gene expression analysis comparing no benefit (NB; n = 22) and clinical benefit (CB; n = 13) groups identified 457 genes upregulated in NB samples (Shukla et al Cell 2018)
Tumor-specific neoantigens are identified using algorithms to predict which mutated peptides are likely to bind to patient-specific MHC class I molecules. TIGL has refined these prediction algorithms incorporating mass spectrometry-based rules as described in the Detection of antigen-specific immunity section. Analysis of large-scale Whole-Exome Sequencing (WES) data using this pipeline has led to numerous insights regarding the drivers of immune responses across cancer and treatment settings.
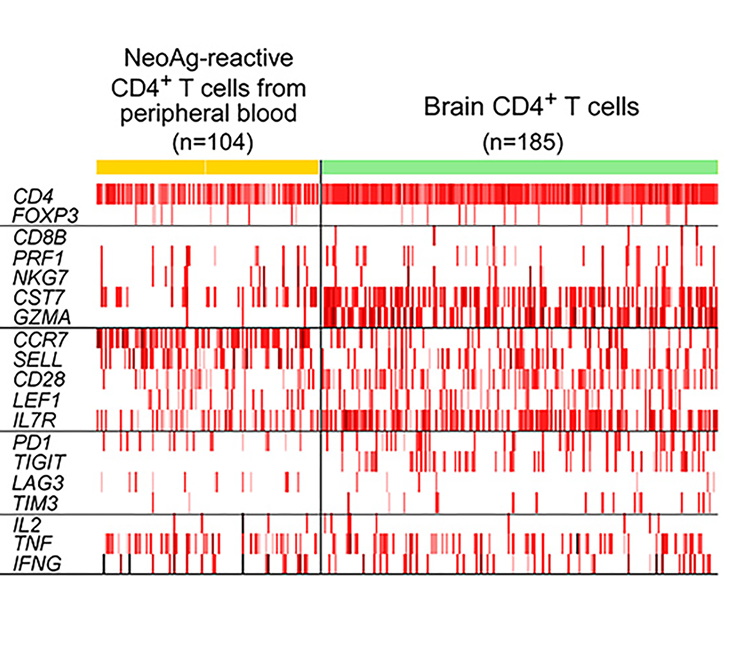
Single cell transcriptome analysis of CD4+ tumor-associated T cells (n=185), freshly isolated at the time of relapse and neoantigen-reactive CD4+ cells isolated from post-vaccination (week 16) peripheral blood (n=104) in glioblastoma Patient 7 (excluding CD4 dropouts). Tumor-associated T cells expressed more granzyme A than circulating neoantigen-reactive CD4+ cells, and showed higher expression of the co-inhibitory molecule TIGIT (Keskin et al Nature 2018)
Staff
Jeremy Simon, PhD, Computational Lead Scientist
Chloe Tu, Computational Biologist
Cleo Forman, Computational Biologist
Bohoon Shim, Associate Computational Biologist
Yiren Shao, Computational Biologist
Angie McGraw, Computational Biologist